-Search query
-Search result
Showing 1 - 50 of 67 items for (author: stansfeld & pj)

EMDB-16489: 
In situ structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-16492: 
In situ structure of the Nitrosopumilus maritimus S-layer - Composite map between C2 and C6
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-16482: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)
Method: single particle / : von Kuegelgen A, Bharat T

EMDB-16483: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)
Method: single particle / : von Kuegelgen A, Bharat T

EMDB-16484: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Composite map between two and six-fold symmetrised
Method: single particle / : von Kuegelgen A, Bharat T

EMDB-16486: 
In vitro Nitrosopumilus maritimus S-layer with NH4Cl
Method: single particle / : von Kuegelgen A, van Dorst S, Bharat TAM

EMDB-16487: 
In situ structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-16657: 
Cryo-EM structure of the fd bacteriophage capsid major coat protein pVIII
Method: helical / : Boehning J, Bharat TAM
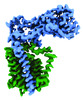
EMDB-41299: 
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Method: single particle / : Nygaard R, Mancia F
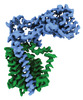
EMDB-41303: 
Transmembrane map
Method: single particle / : Nygaard R, Mancia F
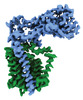
EMDB-41304: 
Periplasmic map
Method: single particle / : Nygaard R, Mancia F

EMDB-15779: 
CryoEM structure of C5b8-CD59
Method: single particle / : Bubeck D, Couves EC, Gardner S

EMDB-15780: 
2C9, C5b9-CD59 structure
Method: single particle / : Couves EC, Gardner S, Bubeck D

EMDB-15781: 
2C9, C5b9-CD59 cryoEM structure
Method: single particle / : Couves EC, Gardner S, Bubeck D

EMDB-15782: 
2C9, C5b9-CD59 cryoEM structure; focus refinement map
Method: single particle / : Couves EC, Gardner S, Bubeck D

EMDB-15783: 
3C9, C5b9-CD59 cryoEM structure; focus refinement map
Method: single particle / : Couves EC, Gardner S, Bubeck D

EMDB-15800: 
CryoEM reconstruction of C5b8-CD59, density subtracted
Method: single particle / : Bubeck D, Couves EC, Gardner S

EMDB-15641: 
Structure of the Native Chemotaxis Core Signalling Complex from E-gene lysed E. coli cells.
Method: subtomogram averaging / : Qin Z, Zhang P

EMDB-15642: 
Native Chemotaxis Core Signalling Complex from E. coli, with focused alignment on the baseplate (CheA-CheW)
Method: subtomogram averaging / : Qin Z, Zhang P

EMDB-15643: 
Native Chemotaxis Core Signalling Complex from E. coli, Focused alignment on ligand binding domain
Method: subtomogram averaging / : Qin Z, Zhang P

EMDB-15894: 
KimA from B. subtilis with nucleotide second-messenger c-di-AMP bound
Method: single particle / : Vonck J, Wieferig JP

EMDB-15895: 
Upright KimA dimer with bound c-di-AMP from B. subtilis
Method: single particle / : Vonck J, Wieferig JP

EMDB-15669: 
Jumbo Phage phi-kp24 tail outer sheath
Method: helical / : Ouyang R, Briegel A

EMDB-14356: 
Jumbo Phage phi-Kp24 full capsid
Method: single particle / : Ouyang R, Briegel A

EMDB-14357: 
Jumbo Phage phi-Kp24 extended tail
Method: single particle / : Ouyang R, Briegel A

EMDB-13862: 
Jumbo Phage phi-Kp24 empty capsid
Method: single particle / : Ouyang R, Briegel A

EMDB-14347: 
Cryo-EM map of the WT KdpFABC complex in the E2-P conformation, stabilised with the inhibitor orthovanadate
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14911: 
Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14912: 
Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Dubach VRA, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14913: 
Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Rheinberger J, Wunnicke D, Dubach VRA, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14914: 
Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14915: 
Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14916: 
Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14917: 
Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Dubach VRA, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14918: 
Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Dubach VRA, Stansfeld PJ, Haenelt I, Paulino C

EMDB-14919: 
Cryo-EM map of the unphosphorylated KdpFABC complex in the E1-P_ADP conformation, under turnover conditions
Method: single particle / : Hielkema L, Stock C, Silberberg JM, Corey RA, Wunnicke D, Dubach VRA, Stansfeld PJ, Haenelt I, Paulino C
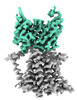
EMDB-26054: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
EMDB-26057: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

EMDB-25366: 
Chlorella virus hyaluronan synthase
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25367: 
Chlorella virus hyaluronan synthase inhibited by UDP
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25368: 
Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25369: 
Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25370: 
Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-13355: 
Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium
Method: single particle / : von Kugelgen A, Bharat TAM

EMDB-13103: 
Cryo-EM structure of yeast Sei1
Method: single particle / : Deme JC, Lea SM

EMDB-13104: 
Cryo-EM structure of yeast Sei1 with locking helix deletion
Method: single particle / : Deme JC, Lea SM

EMDB-12478: 
Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+
Method: single particle / : Silberberg JM, Corey RA, Hielkema L, Stock C, Stansfeld PJ, Paulino C, Haenelt I

EMDB-12482: 
Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex.
Method: single particle / : Silberberg JM, Corey RA, Hielkema L, Stock C, Stansfeld PJ, Paulino C, Haenelt I

EMDB-22843: 
Cryo-electron microscopy structure of the heavy metal efflux pump CusA in the symmetric closed state
Method: single particle / : Moseng MA

EMDB-22844: 
Cryo-electron microscopy structure of the heavy metal efflux pump CusA in a homogeneous binding copper(1) state
Method: single particle / : Moseng MA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
